Did life start in a hot spring? Study tracing ancestor microorganisms suggests so
Earth's environment for first organisms living four billion years ago was very different to how it is today.

Jeff Errington, Newcastle University
It's one of the greatest mysteries of modern science: how did life begin exactly? While most scientists believe that all lifeforms evolved from a common, primitive ancestor microorganism, the details are blurry. What kinds of genes did this lifeform carry and where did it live? A new study, published in Nature Microbiology, now sheds some light on this early organism and the environment it evolved in.
Experimental scientists interested in the origins of life generally tackle the problem in two distinct ways. One is a bottom-up approach in which they try to imagine how early life might have emerged and then try to recreate the key steps in the laboratory. The alternative, a top-down approach, is to analyse or strip down modern cells to simplify them and deduce how the key stages in the evolution of complexity might have taken place.
Informaticians interested in this problem exploit the huge amounts of data emerging from the revolution in DNA sequencing. This has resulted in a sea of information about the genomes of organisms – from bacteria to humans. Hidden in this should be the echoes of DNA sequences from primitive cells – the first cells on the planet to use the modern genetic code – passed on through billions of generations.
The "last universal common ancestor" is a hypothetical very early single cell from which all life on Earth descended. The relationship between this ancestor and modern organisms is often visualised in the form of evolutionary trees, of which the most famous early examples were those by Charles Darwin.
The advent of DNA sequencing provided a wonderful, highly quantitative measure of genetic relatedness that transcended the whole of biology. The same four-base code of A, C, G and T is used by virtually all organisms on the planet. So in principle, it can be used to construct evolutionary trees for the whole of life. We know that certain genes, such as the one encoding a small RNA subunit of the ribosome (the protein synthesisers of a cell), existed at the dawn of cellular life on Earth and seem to have been inherited by all subsequent forms of life. Over 4 billion years, copies of this particular gene – 16S rRNA – have gradually changed by random mutation in the separate lineages that have led to different forms of life. This means each has a characteristic sequence that is similar in recently diverged organisms but increasingly different in lineages that diverged earlier in evolution.
The first analyses of these "universal" DNA sequences about 30 years ago led to dramatic changes in our appreciation of the diversity of life on Earth, and especially the staggering degree of diversity in single celled organisms with no nucleus (the prokaryotes). It also highlighted the existence of a huge new "domain" of prokarytic life – now called the archaea.
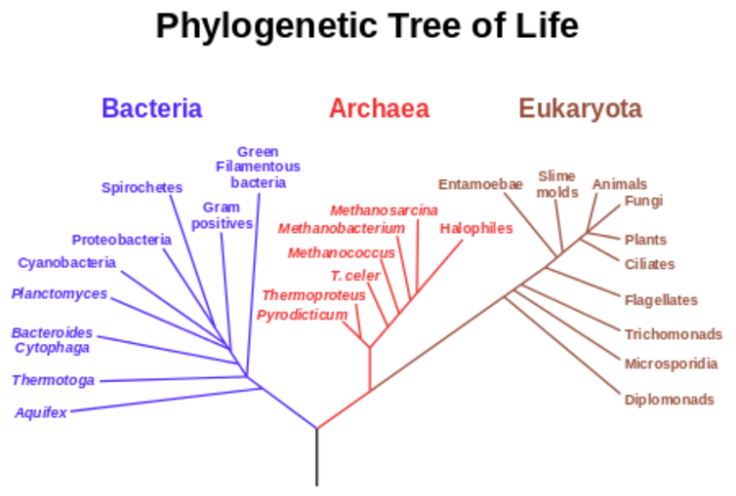
Attempts to develop truly universal trees that would define how all modern cells descended from this last universal ancestor have been thwarted by a number of technical issues. One problem lies in the sheer number of groups that have separated from each other since life first began. What's more, bacteria can also exchange genes with each other, which makes it harder to identify how they've been passed down.
Hydrogen-powered organism?
The researchers behind the new study applied a sophisticated state-of-the-art method to organise some 6m sequenced prokaryotic genes into families. They then looked for patterns of similarity across all bacterial groups and found a small set of genes that were present in both archaea and bacteria. They could show that these genes were really likely to have been inherited directly from a common ancestor rather than by lateral exchange along the way.
The result is important because it identifies specific groups of bacteria (clostridia) and archaea (methanogens) that carry early versions of these genes, meaning they are very ancient and may be similar to the very earliest organisms that gave rise to the separate bacterial and archaeal lineages.
More importantly, the nature of the genes that are conserved tells an amazing story about the kind of environment in which this last common ancestor lived – including how it extracted energy to survive and thrive. The study suggests that the world inhabited by these organisms nearly four billion years ago was very different to the one we live in now. There was no available oxygen, but according to the genes, this common ancestor probably obtained energy from hydrogen gas, presumably made by geochemical activity in the Earth's crust.
"Inert" gases including carbon dioxide and nitrogen would have provided the key building blocks for making all cellular structures. Iron was freely available, with no oxygen to turn it into insoluble rust, and so this element was used by many enzymes in the early cell. Some of the genes are believed to be involved in adaptation to high temperatures, which suggests these organisms evolved in a hydrothermal environment – perhaps equivalent to modern hydrothermal vents or hot springs, where some bacteria still thrive.
Sadly, without a time machine, there is no way to directly verify these results. Nevertheless, this information will now be of great interest, not least to those scientists wishing to use the information to inform their bottom-up experiments in recreating modern forms of primitive life. But it will not be easy, given the requirement for high temperature, nitrogen, carbon dioxide and explosive hydrogen gas.
Jeff Errington, Director of the Centre for Bacterial Cell Biology, Newcastle University
This article was originally published on The Conversation. Read the original article.
© Copyright IBTimes 2025. All rights reserved.





















